Mixing it up - From Transposons to GENE Expression systems
I think it is important that we expand a little on the genetic tools available on Drosophila flies. Not only to know more about these tools, but also to gaze into the wonders of our genomes. The DNA we have in every cell of our bodies is not a static, fixed molecule. It is subject to changes. Some parts of it can change place and some pieces can be incorporated. Transgenesis is a process that has been occurring naturally for ages.
We will also look at this in a bit more of a historical perspective, to see how the power of these basic science discoveries was harnessed to translate them into techniques and tools (such as gene expression systems) that revolutionized the use of Drosophila as a model organism and paved the way for decades of research and discoveries.
Transposable elements - P Elements
In the post about transgenic flies, we did not give any details on how the engineered DNA we put into a plasmid (a circular molecule of DNA that can be replicated in bacteria) gets afterwards into the genome of the fly. We mentioned briefly that this happens because we use an engineered plasmid that has some DNA sequences that are recognized by a protein that can cut them and then move it and paste it somewhere in the genome at random. This protein is called a transposase because it can transpose pieces of DNA within the genome.
In the 1950s Barbara McClintock observed specific patterns of pigment abnormalities in maize plants. She discovered that the genome is not static and that some DNA in it can move around and change place. When these pieces of DNA move sometimes they land on genes and interrupt their function, generating mutations such as the loss of the capacity to produce some pigments.
Because they can change position in the genome, these sequences are called transposable elements or transposons (McClintock work went unnoticed and disregarded at the time; it was later rediscovered and she was awarded the Nobel Prize much later).
In fact, we have mentioned that a lot of the DNA in our genomes does not contain actual genes, well a huge part of all that extra DNA in all the cells of our own human bodies are the remains of elements that moved around and left pieces behind through the millions of years of evolution. Our genomes are a true DNA patchwork!
These DNA elements were later found to be specific sequences that use different mechanisms to move around the genome.
Some of them move in two steps; first the transcription of the DNA sequence into RNA (as we already learned) and second a process of turning that RNA back into DNA (called retrotranscription) to finally insert it somewhere in the genome (viruses such as HIV work this way).
Others change place through the cutting of the actual piece of DNA from the genome to then transport it and insert it somewhere else in the genome, a process mediated by the transposase.
Some elements are autonomous and the processes to move them around are carried out by proteins encoded within the elements themselves. They can move on their own!
In the 1960s and 1970s transposable elements were intensely studied and found in many organisms. At the end of the 1970s and beginning of the 1980s they were proposed as the explanation for a very interesting phenomenon observed in Drosophila flies than involved unexpected mutations. This led to the discovery of transposable elements called P elements.
The transposable elements found (the P elements) consisted of a gene that codes to produce a transposase protein. At both ends of this gene are the sequences that the transposase can bind and then cut, to take the whole piece (including the transposase gene) out from the genome and insert it somewhere else.
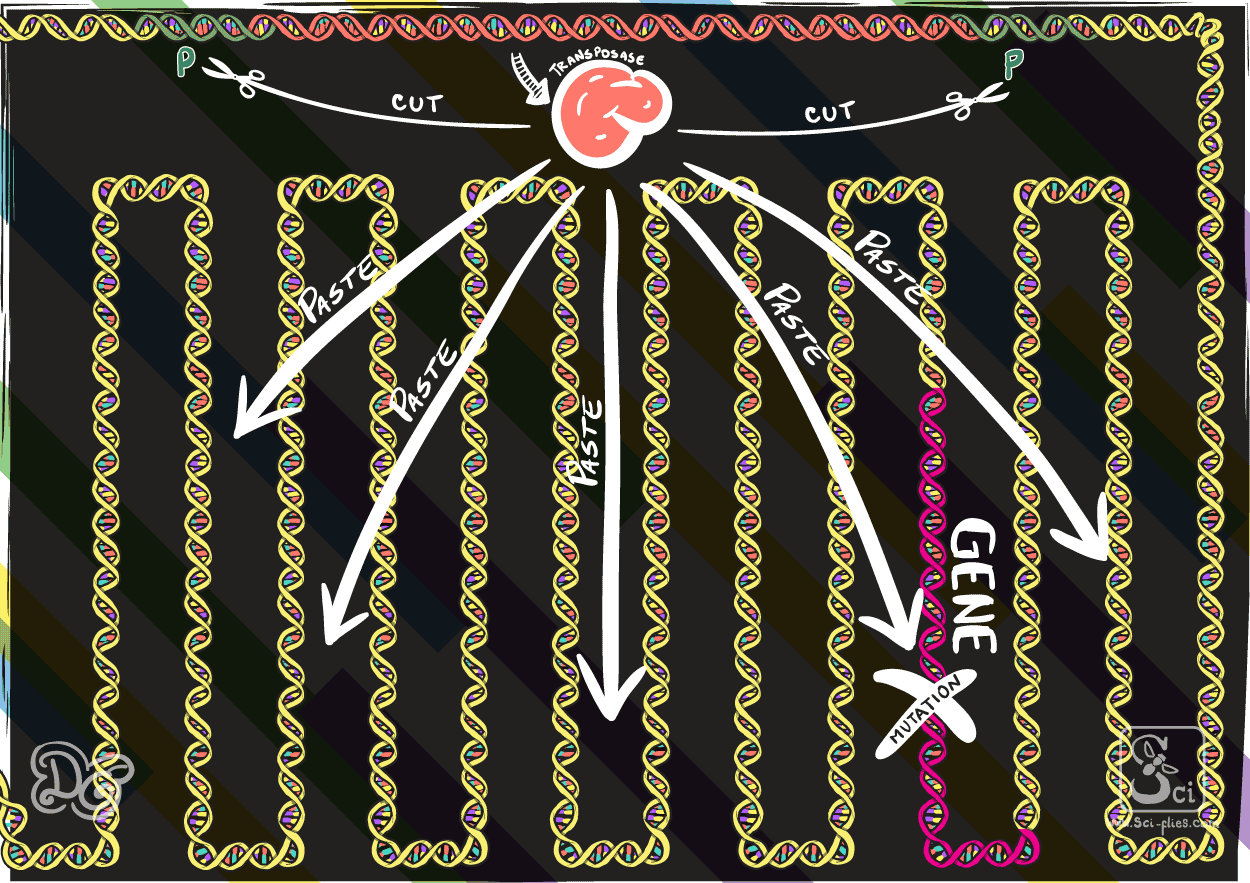
In 1982 Gerald Rubin and Allan Spradling injected these elements into embryos of fly strains that did not normally have them and showed that in the next generations those elements were incorporated in the genome and could move around the genome.
They realized that they could use these elements that occurred naturally in the flies to insert any piece of DNA in the genome by replacing the transposase in the element with the DNA they wanted to insert. In this way they proved the concept to make transgenic flies through embryo injection as we already described.
Transposable elements were and are interesting in their own right as they are elements that can move around the genome and cause mutations. How do they work? How do they move? Can they be lethal? How are they regulated to not be lethal? Why are they still around in the flies if they can cause bad mutations? Many questions that are still being studied.
And those very interesting questions also resulted in a powerful invention, the way to put any gene from any source inside the fly, a tool for further discoveries that revolutionized the use of Drosophila as a model organism.
How did this continue?
More uses of transposable elements. Enhancer traps.
In the post about transgenic flies we said that some DNA that does not contain information to make RNA or proteins, contains the instructions to set up the machinery that is in charge of using the information in DNA to make RNA (the transcription machinery). Those regions are located around the gene they control and are called promoters and enhancers.
Promoters are the ones that contain the minimal information to setup of the machinery while enhancers are the ones that tell about the time and place where expression should take place (we call expression to the transcription of a gene, the production of RNA from DNA).
We used as a fictional example the idea of copying the instructions for the expression of a gene and using them to tell the transcription machinery to produce the RNA for a fluorescent protein (GFP). In this way we can see when in the life cycle and where in the fly body the gene is normally expressed by looking at the fluorescence with a microscope.
But, what happens if we do not know which genes are involved in the biological process of our interest? How can we discover those genes?
At the end of the 1980s the laboratory of Walter Gehring made a tool to discover these regions that contain instructions for expression. This tool is called enhancer trap and uses the P elements and the transposase we mentioned before in a very creative way.
They constructed a plasmid with the P elements around a gene to make a protein that could be visualized. Let’s call this a reporter because it can report the expression of other genes. They made a transgenic fly that had this reporter.
On the other hand, the laboratories of Engels, Rubin and Spradling had generated tools to move inserted P elements around (we will talk further about this when we talk about mutants in the future). This specific tool is a fly that has the transposase gene fixed in the genome, unable to jump around.
They crossed these two lines to have a progeny that had both elements (remember Mendel?), the transposase and the reporter gene. Now, the transposase can cut the DNA around P elements and insert it randomly in the genome. The reporter was around P elements so in the progeny of the flies that had these two elements, the reporter would be moved from its original place into a random location in the genome.
Flies produce a lot of eggs and every one of those offspring would have the reporter in a different location. By chance, a proportion of these random insertions would be located next to sequences with the instructions for the expression of different genes.
By later visualizing the reporter they could discover genes that had interesting expression. These were called enhancer trap strains, because they had used a random reporter to trap the enhancers of genes.
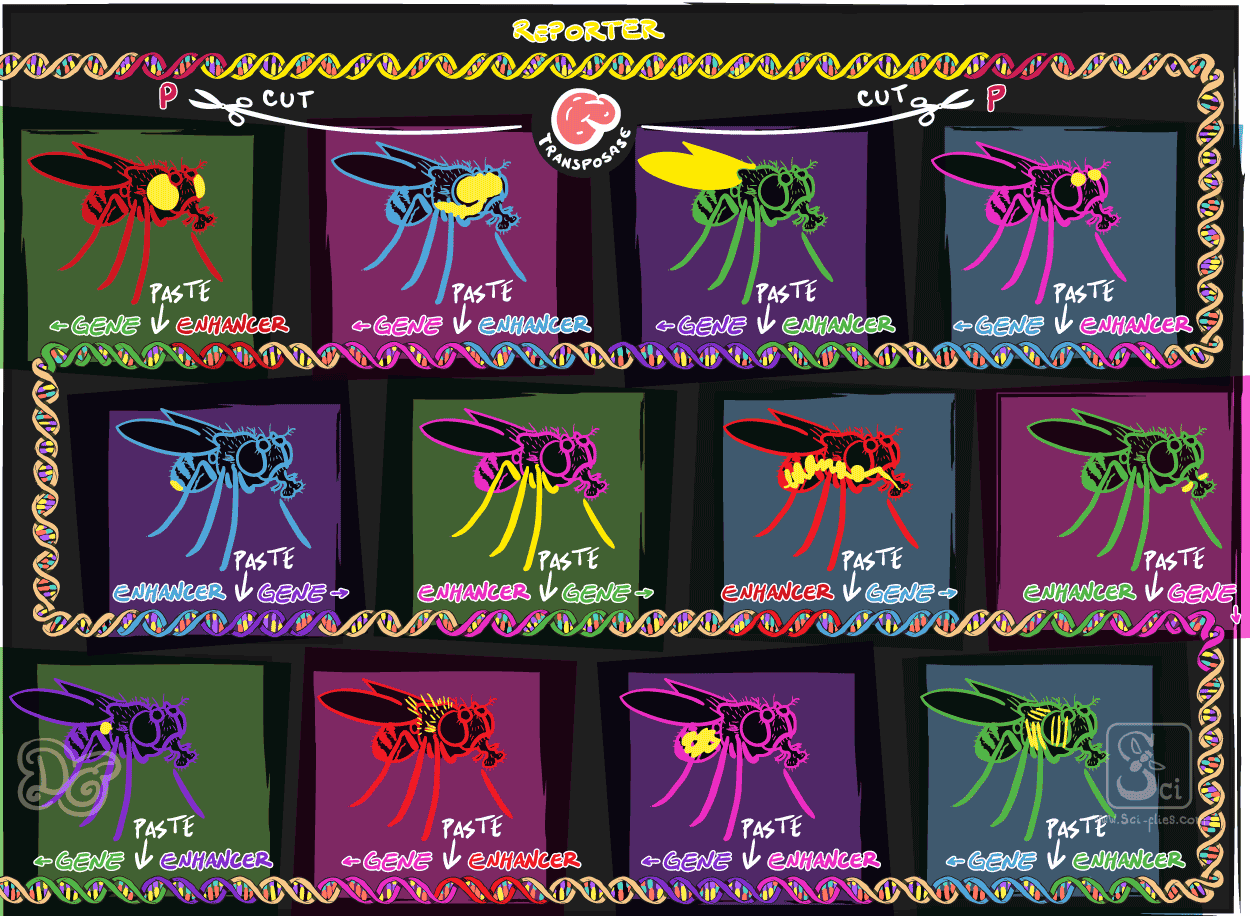
Later in history this strategy was modified to use GFP or other genes as reporters. Huge collections of these lines were produced, and many interesting genes were discovered in this way.
One interesting use of these enhancer trap lines is to be able to observe how expression of genes is affected by modifications to the fly’s environment. For example, genes that change expression when flies are fed different food, or exposed to different temperatures, etc.
What else can you think of?
With better understanding of how enhancers work, it was discovered that pieces of the sequences of enhancers could be used to tell the machinery to express a gene in more specific places, sometimes even in one or two specific cells. Huge collections of these transgenic strains exist for anyone to use.
The Gal4-UAS expression system
What if we want to induce expression of some gene?
In the post about transgenic flies we talked about putting the sequence with the instructions in a plasmid in front of the gene we want to express and making a transgenic fly. But what if we want to express that same gene in another way? Well then we would have to make another transgenic fly with different instructions. And a new one for every different expression we want to induce. That is very time consuming and not very practical.
So, what do we do? How do we avoid having to make a new transgenic strain for every combination?
The answer was developed in the mid-1990s by Andrea Brand and Norbert Perrimon. They had the brilliant insight of separating the two elements of the system and having the instructing sequences on one fly and the gene of interest to express on another. When those flies are crossed, the offspring carry both parts and expression is achieved (always remember Mendel!).
But how? How are the two elements connected? The instructions to express a gene need to be next to the gene, or not?
The solution was to use an intermediary, an adaptor between the two parts. In this case, the fly’s instruction sequences will control the expression of a gene from yeasts, called Gal4. This gene does not exist in flies and codes to produce a transcription factor.
As we mentioned before, transcription factors are the proteins that read the instructions to provoke transcription (they bind to promoters and enhancers) and activate expression of genes. The Gal4 transcription factor recognizes specific instruction sequences from yeasts that also do not exist in flies called UAS. This yeast instructions are put in front of our gene of interest.
A somewhat customizable control of expression could be achieved by using another type of controlling sequence.
There is a protein called the heat shock factor, this protein is produced when the temperature of the organism rises above a dangerous level. This protein is part of a mechanism to protect the cells from the destructive effects of high temperature.
Using the enhancer region of the heat shock gene we can provoke expression of any other gene giving the animal a short exposure to high temperature (for a fly 37C – 98.6F is high, their optimal temperature for living is 25C – 77F).
In this way we can control the moment at which the gene will be expressed (the timing of expression) through the heat pulse.
However, we cannot control where in the body of the fly it will be expressed because it is in every cell of the fly.
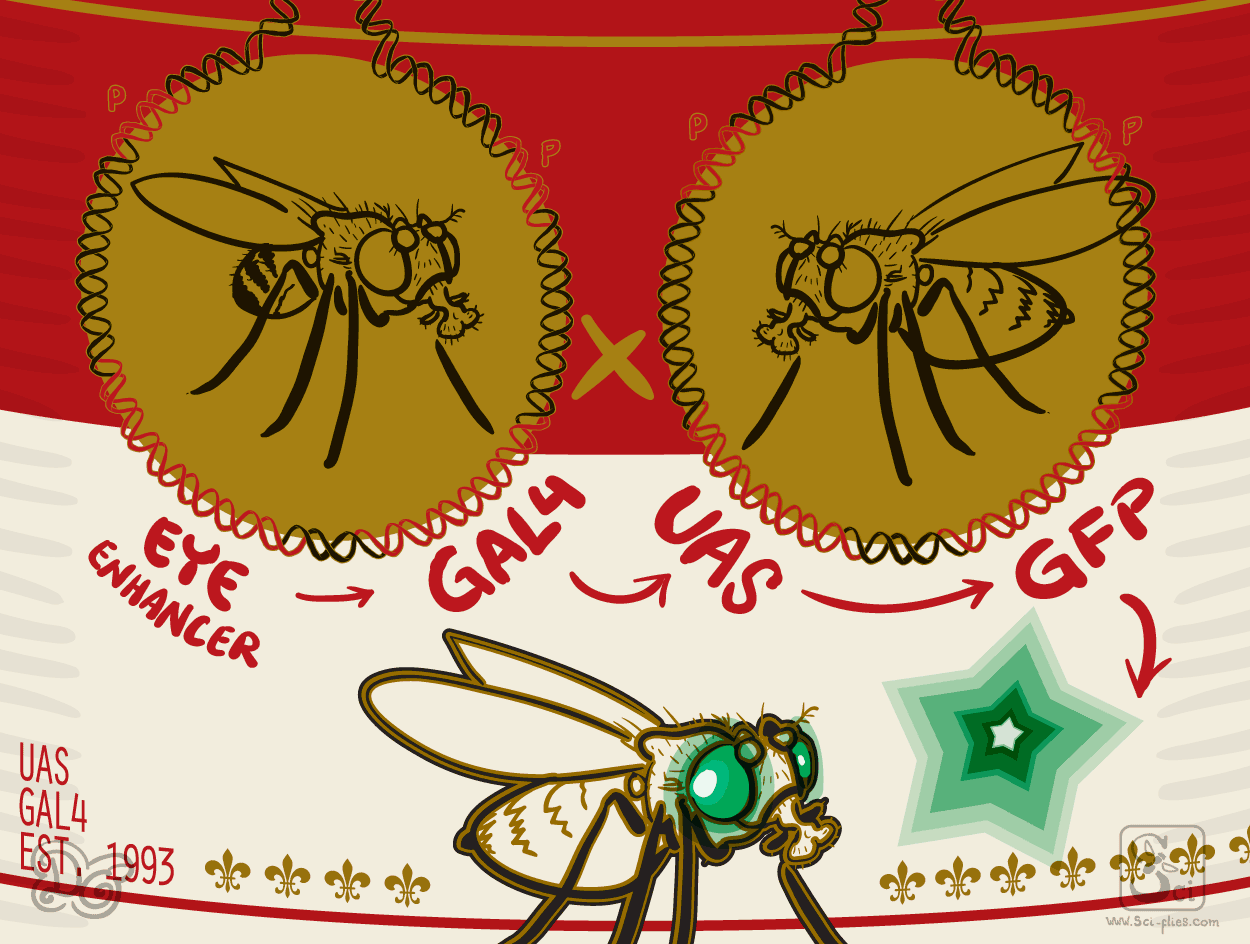
Now we have a two-part system. Enhancer-Gal4 flies with instructions to produce Gal4 in a specific time and place and UAS-Gene of interest, that will be activated only when and where Gal4 is present. Enhancer-Gal4 flies can be crossed to UAS-Gene of interest flies and produce offspring that have both elements.
This gave an incredible versatility to fly transgenics. Brand and Perrimon used the same enhancer trap approach to generate a collection of random Gal4 insertions. They made an original transgenic fly with a Gal4 gene around P elements and crossed it to the transposase line. In the offspring the Gal4 gene was moved around to random places. Some places were next to enhancers, which induced the production of Gal4 protein in the expression pattern of unknown genes.
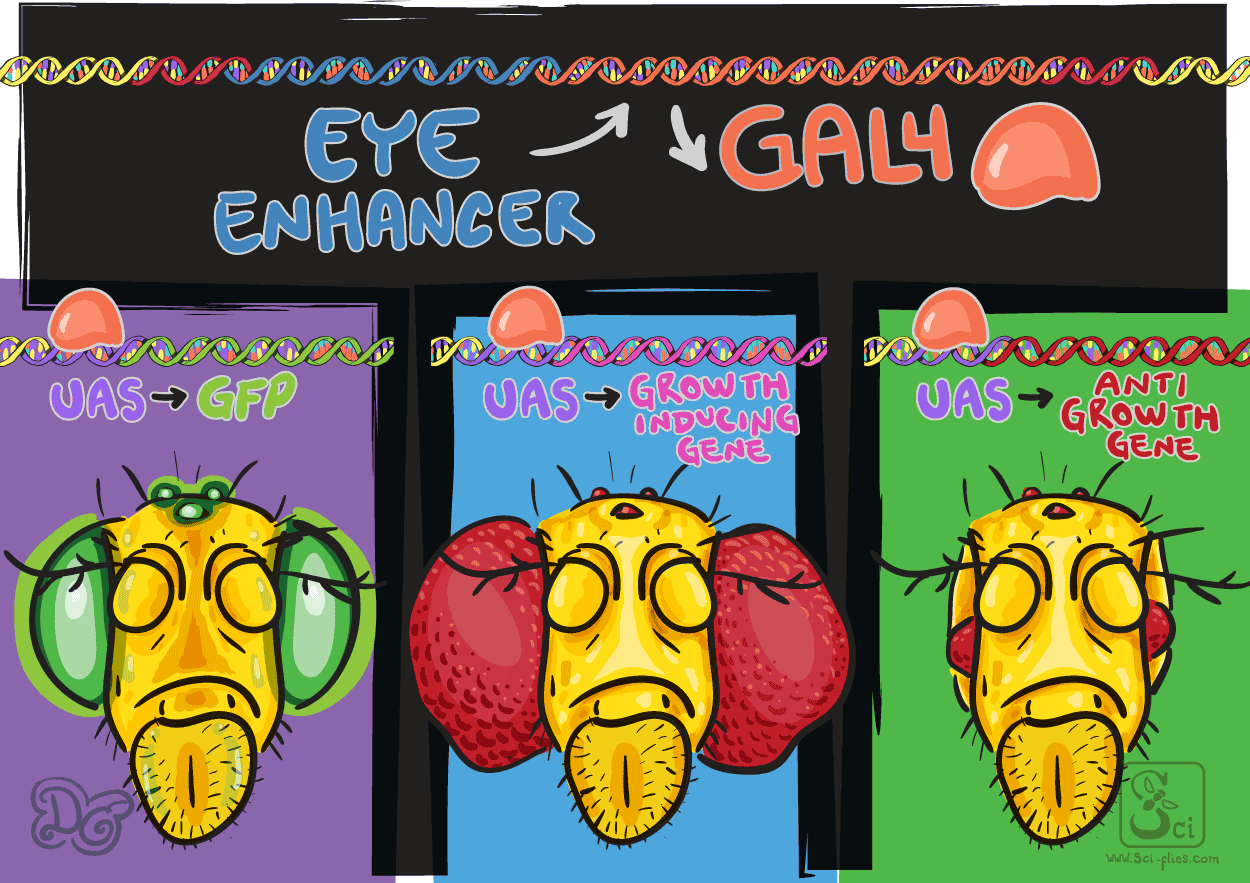
Suddenly any gene could be expressed in potentially any place and time. UAS transgenic flies could be made to carry reporter genes (such as GFP) or genes that have a specific function. Also, genes from other species, or modified genes with missing parts or changed codons to alter their protein products and understand how their function relates to their structure.
For example, it became possible to kill off specific cells, by expressing genes that induce cell death, and see what happens when their function is lost. Or to express variants of proteins that function as repressors. Or to express developmental genes in a different pattern to better understand how their function is related to the time and place of their expression.
And many, many, more things. We will certainly expand on these later.
The generation of these tools from the beginning of the 1980s to the mid-1990s constituted a revolution in the use of Drosophila flies as a malleable, genetic model organism. The way of thinking experiments changed and the questions that could be addressed also expanded. Now it was possible to dissect the molecular, genetic and cellular components of almost all aspects of fly biology.
The description of transposable elements in the fly, led to the understanding of their molecular mechanisms, which in turn allowed their manipulation and technical use. As new molecular mechanisms are constantly being discovered and described, their manipulation also becomes possible, and in this way new tools are continually being developed. We will continue to talk about interesting tools and techniques in the future.
In the next post we will talk about some strategies for discovering interesting genes and a very important historical example.
Thanks for reading and see you again soon!
Interesting References
I based most of the text on the original papers by Rubin, Bingham, Kidwell; Rubin and Spradling; Bellen, O’Kane, Willson, Grossniklaus, Pearson and Gehring; Brand and Perrimon; and some others. I had a lot of fun reading those papers. I also took from an interview to Rubin in (Dis. Model Mech. 2016, vol. 9 (4) pp 361-364). I have also used a recent review by Halles et al. on Drosophila techniques (Genetics, 2015 vol. 201, 3 pp 815-842) that I recommend for everyone interested.